Publications
2025
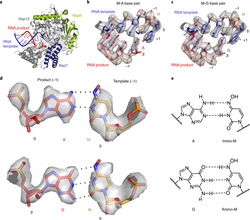
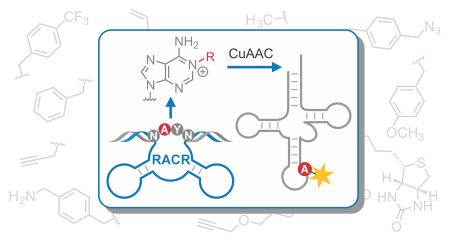
C. P. M. Scheitl, E. Dorinova, S. Christopher, M. B. Walunj, T. Okuda, M. V. Sednev, C. Höbartner, An Unexpected Adenosine-Alkylating Ribozyme Emerged by Target Site Relocation during in Vitro Selection. Journal of the American Chemical Society 2025, published online November 20, https://doi.org/10.1021/jacs.5c13970
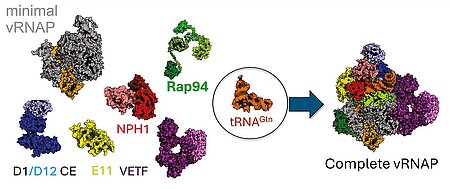
J. Bartuli, S. Jungwirth, M. Dixit, T. Okuda, J. P. Zimmermann, M. Erlacher, T. Pan, A. Volz, A. Hüttenhofer, B. Warscheid, C. Höbartner, C. Grimm, U. Fischer, "tRNA as an assembly chaperone for a macromolecular transcription-processing complex", Nature Structural & Molecular Biology 2025, published online September 4, https://doi.org/10.1038/s41594-025-01653-y
R. C. Kretsch, R. Albrecht, E. S. Andersen, H.-A. Chen, W. Chiu, R. Das, J. G. Gezelle, M. D. Hartmann, C. Höbartner, Y. Hu, S. Jadhav, P. E. Johnson, C. P. Jones, D. Koirala, E. L. Kristoffersen, E. Largy, A. Lewicka, C. D. Mackereth, M. Marcia, M. Nigro, M. Ojha, J. A. Piccirilli, P. A. Rice, H. Shin, A.-L. Steckelberg, Z. Su, Y. Srivastava, L. Wang, Y. Wu, J. Xie, N. H. Zwergius, J. Moult, A. Kryshtafovych, "Functional Relevance of CASP16 Nucleic Acid Predictions as Evaluated by Structure Providers", Proteins – Structure | Function | Bioinformatics 2025, published online September 4, https://doi.org/10.1002/prot.70043
R. C. Kretsch, R. Albrecht, E. S. Andersen, H.-A. Chen, W. Chiu, R. Das, Jeanine G. Gezelle, Marcus D. Hartmann, Claudia Höbartner, Y. Hu, S. Jadhav, P. E. Johnson, C. P. Jones, D. Koirala, E. L. Kristoffersen, Emil L. E. Largy, A. Lewicka, C. D. Mackereth, M. Marcia, M. Nigro, M. Ojha, J. A. Piccirilli, P. A. Rice, H. Shin, A.-L- Steckelberg, Z. Su, Y. Srivastava, L. Wang, Y. Wu, J. Xie, N. H. Zwergius, J. Moult, A. Kryshtafovych, "Functional relevance of CASP16 nucleic acid predictions as evaluated by structure providers", bioRxiv 2025, preprint April 22, https://doi.org/10.1101/2025.04.15.649049
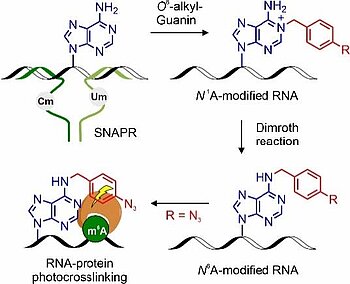
M. B. Walunj, C. P. M. Scheitl, T. Jungnickel, C. Höbartner. "Ribozyme-catalyzed site-specific labeling of RNA using O6-alkylguanine SNAP-tag substrates". Angew. Chem. Int. Ed 2025, first published: April 15, https://doi.org/10.1002/anie.202500257
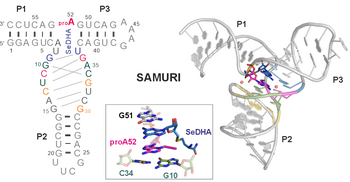
H.-A. Chen, T. Okuda, A-K. Lenz, C. P. M. Scheitl, H. Schindelin, C. Höbartner. "Structure and catalytic activity of the SAM utilizing ribozyme SAMURI" Nature Chemical Biology 2025, published online January 8, https://doi.org/10.1038/s41589-024-01808-w
2024
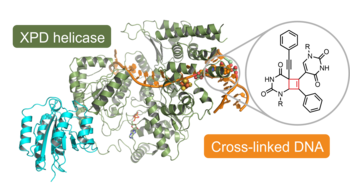
J. Kuper, T. Hove, S. Maidl, H. Neitz, F. Sauer, M. Kempf, T. Schroeder, E. Greiter, C. Höbartner & C. Kisker. "XPD stalled on cross-linked DNA provides insight into damage verification", Nature Structural and Molecular Biology 2024, 31, pages1580–1588, https://doi.org/10.1038/s41594-024-01323-5
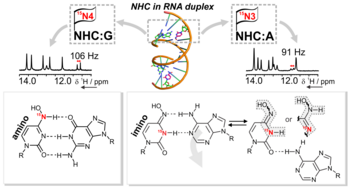
I. Bessi, C. Stiller, T. Schroeder, B. Schäd, M. Grüne, J. Dietzsch, C. Höbartner, The Tautomeric State of N4-Hydroxycytidine within Base-Paired RNA, ACS Cent. Sci. 2024, 10, 5, 1084–1093, https://doi.org/10.1021/acscentsci.4c00146
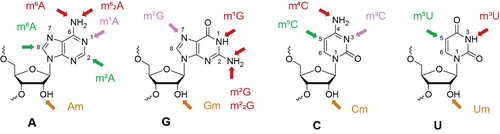
C. Höbartner, K.E. Bohnsack, M.T. Bohnsack. How Natural Enzymes and Synthetic Ribozymes Generate Methylated Nucleotides in RNA. Annu Rev Biochem. 2024, Vol. 93: 109-137, 10.1146/annurev-biochem-030222-112310.
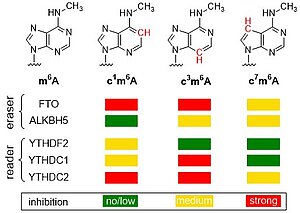
F. Seitz, T. Jungnickel, N. Kleiber, J. Kretschmer, J. Adelmann, J. Dietzsch, K.E. Bohnsack, M.T. Bohnsack, C. Höbartner. Atomic mutagenesis of N6-methyladenosine reveals distinct recognition modes of human m6A reader and eraser proteins J. Am. Chem. Soc. 2024, 146, 11, 7803–7810, doi.org/10.1021/jacs.4c00626
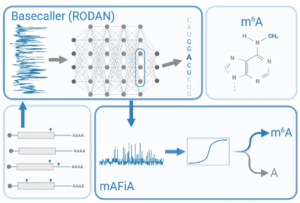
A. Chan, I. S. Naarmann-de Vries, C. P. M. Scheitl, C. Höbartner, C. Dieterich, Detecting m6A at single-molecular resolution via direct-RNA sequencing and realistic training data (mAFiA, bioRxiv 2023, 10.1101/2023.07.28.550944, Nature Communications 15, 2024, 3323. doi.org/10.1038/s41467-024-47661-2
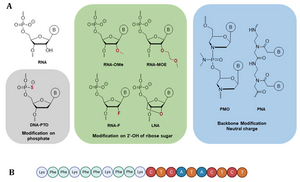
C. Gosh, L.Popella, V. Dhamodharan, J. Jung, J. Dietzsch, L. Barquist, C. Höbartner, J. Vogel. A comparative analysis of peptide-delivered antisense antibiotics using diverse nucleotide mimics, RNA 2024, 30: 624-643, 10.1261/rna.079969.124
J. Kuper, T. Hove, S. Maidl, F. Sauer, M. Kempf, E. Greiter, H. Neitz, C. Höbartner, C. Kisker, Trapped in translocation: Stalling of XPD on a crosslinked DNA substrate, biorxiv 2024.02. 20.581127
M. Müller, H. Neitz, C. Höbartner, H. Helten, BN-Phenanthrene- and BN-Pyrene-Based Fluorescent Uridine Analogues. Org. Lett. 2024, https://doi.org/10.1021/acs.orglett.3c04226
C. Höbartner, Ribozymes and RNA Modifications, in Chemistry Callenges of the 21st Century, The Solvay Institute, 2024, 453-459. https://doi.org/10.1142/9789811282324_0043
2023
D. Fischermeier, C. Steinmetzger, C Höbartner, R. Mitrić. Conformational preferences of modified nucleobases in RNA aptamers and their effect on Förster resonant energy transfer. Phys Chem Chem Phys 2023 Dec 6. doi: 10.1039/d3cp04704k
Helm M, Bohnsack MT, Carell T, Dalpke A, Entian KD, Ehrenhofer-Murray A, Ficner R, Hammann C, Höbartner C, Jäschke A, Jeltsch A, Kaiser S, Klassen R, Leidel SA, Marx A, Mörl M, Meier JC, Meister G, Rentmeister A, Rodnina M, Roignant JY, Schaffrath R, Stadler P, Stafforst T. Experience with German Research Consortia in the Field of Chemical Biology of Native Nucleic Acid Modifications. ACS Chem Biol. 2023 Nov 14. doi: 10.1021/acschembio.3c00586
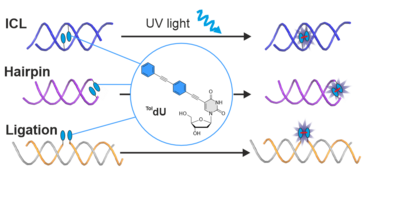
H. Neitz and C. Höbartner. A tolane-modified 5-ethynyluridine as a universal and fluorogenic photochemical DNA crosslinker. Chem Commun. 2023 59, 12003-12006. doi: 10.1039/d3cc03796g.
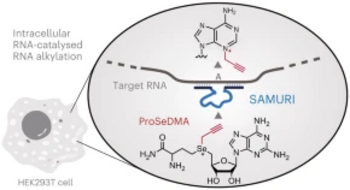
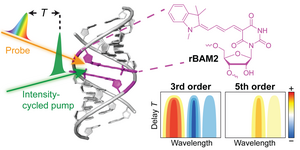
T. Okuda, A.-K. Lenz, F. Seitz, J. Vogel, C. Höbartner. A SAM analogue-utilizing ribozyme for site-specific RNA alkylation in living cells, Nat. Chem. 2023, 15, 1523-1531. Epub 2023 Sep 4 doi.org/10.1038/s41557-023-01320-z
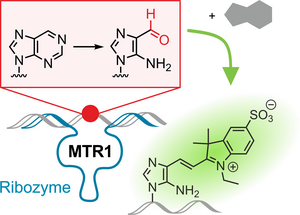
C. P. M. Scheitl, T. Okuda, J. Adelmann, C. Höbartner
Ribozyme-Catalyzed Late-Stage Functionalization and Fluorogenic Labeling of RNA
Angew. Chem. Int. Ed 2023, 62,e202305463. Epub 2023 Jun 23. doi: 10.1002/anie.202305463
J. Dietzsch,
Excitonic coupling of RNA-templated merocyanine dimer studied by higher-order transient absorption spectroscopy
Chem. Commun 2023, 59, 7395-7398. doi: 10.1039/D3CC02024J
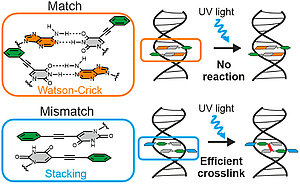
H. Neitz, I. Bessi, J. Kuper, C. Kisker, C. Höbartner
Programmable DNA Interstrand Crosslinking by Alkene–Alkyne [2 + 2] Photocycloaddition
J. Am. Chem. Soc. 2023, 145, 9428–9433. doi: 10.1021/jacs.3c01611
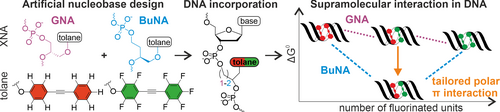
H. Neitz, I. Bessi, V. Kachler, M. Michel, C. Höbartner
Tailored Tolane-Perfluorotolane Assembly as Supramolecular Base Pair Replacement in DNA
Angew. Chem. Int. Ed 2023, 62, e202214456. doi: https://doi.org/10.1002/anie.202214456
2022
C. Steinmetzger, C. Höbartner
Probing of Fluorogenic RNA Aptamers via Supramolecular Förster Resonance Energy Transfer with a Universal Fluorescent Nucleobase Analog
Methods Mol. Biol. 2023, 2570:155-173. First Online: 27 September 2022. doi: https://doi.org/10.1007/978-1-0716-2695-5_12
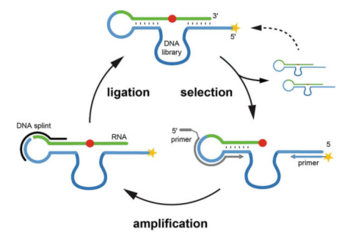
A. Liaqat, M. V. Sednev, C. Höbartner
In Vitro Selection of Deoxyribozymes for the Detection of RNA Modifications
Methods Mol Biol 2022, 2533, 167-179, published July 8, 2022. doi: 10.1007/978-1-0716-2501-9_10
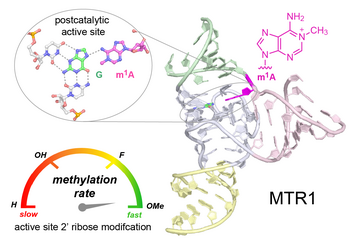
C.P.M. Scheitl, M. Mieczkowski, H. Schindelin, C. Höbartner
Structure and mechanism of the methyltransferase ribozyme MTR1
Nat Chem Biol 2022, 18(5): 547-555, published March 17, 2022. doi: 10.1038/s41589-022-00976-x
B. Liu, Y. Vonhausen, A. Schulz, C. Höbartner, F. Würthner
Peptide Backbone Directed Self-Assembly of Merocyanine Oligomers into Duplex Structures
Angew. Chem. Int. Ed 2022, first published: February 22nd. doi: doi.org/10.1002/anie.202200120
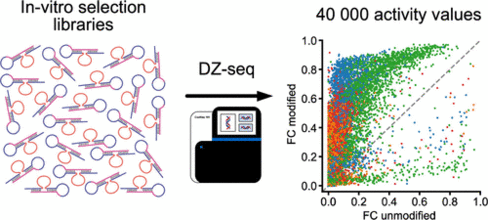
M. V. Sednev, A. Liaqat, C. Höbartner
High-Throughput Activity Profiling of RNA-Cleaving DNA Catalysts by Deoxyribozyme Sequencing (DZ-seq)
J. Am. Chem. Soc. 2022 144, 2090-2094. First published 26 January 2022. doi: doi.org/10.1021/jacs.1c12489
N. Kleiber, N. Lemus-Diaz, C. Stiller, M. Heinrichs, M. Mong-Quyen Mai, P. Hackert, R. Richter-Dennerlein, C. Höbartner, K. E. Bohnsack, M. T. Bohnsack
The RNA methyltransferase METTL8 installs m3C32 in mitochondrial tRNAsThr/Ser(UCN) to optimise tRNA structure and mitochondrial translation
Nat. Commun. 2022, 13, 209. First published 11 January 2022. doi: doi.org/10.1038/s41467-021-27905-1
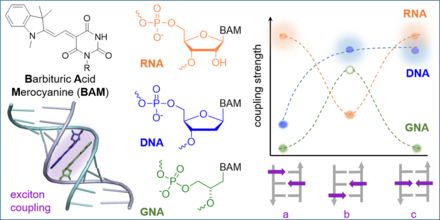
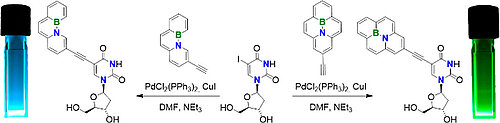
J. Dietzsch, D. Bialas, J. Bandorf, F. Würthner, C. Höbartner
Tuning exciton coupling of merocyanine nucleoside dimers by RNA, DNA and GNA double helix conformations
Angew. Chem. Int. Ed. 2022. doi: 10.1002/anie.202116783
2021
F. Kabinger, C. Stiller, J. Schmitzová, C. Dienemann, G. Kokic, H.S. Hillen, C. Höbartner, P. Cramer
Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis
Nat. Struct. Mol. Biol. 2021, 28, 740-746. First published 11 August 2021. doi: doi.org/10.1038/s41594-021-00651-0
A. Liaqat, M.V. Sednev, C. Stiller, C. Höbartner
RNA-cleaving deoxyribozymes differentiate methylated cytidine isomers in RNA
Angew. Chem. Int. Ed. 2021, 60, 19058–19062. First published: 29 June 2021. doi: doi.org/10.1002/anie.202106517 SI
M. Mieczkowski, C. Steinmetzger, I. Bessi, A.-K. Lenz, A. Schmiedel, M. Holzapfel, C. Lambert, V. Pena, C. Höbartner
Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine
Nat. Commun. 2021, 12, 3549. First published 10 June 2021. SI
P. Cramer, G. Kokic, C. Dienemann, C. Höbartner, H.S. Hillen
Coronavirus-Replikation: Mechanismus und Inhibition durch Remdesivir
Biospektrum (Heidelb.) 2021, 27, 49-53. First published (Epub) 12 February 2021. doi: 10.1007/s12268-021-1516-6
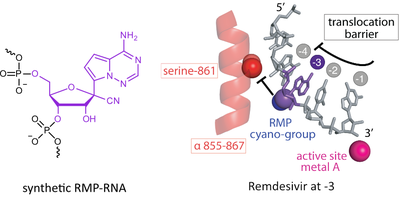
G. Kokic, H.S. Hillen, D. Tegunov, C. Dienemann, F. Seitz, J. Schmitzova, L. Farnung, A. Siewert, C. Höbartner, P. Cramer
Mechanism of SARS-CoV-2 polymerase stalling by remdesivir
Nat. Commun. 2021, 12, 279. First published 12 January 2021. doi: doi.org/10.1038/s41467-020-20542-0 SI
2020
S. Werner, A. Galliot, F. Pichot, T. Kemmer, V. Marchand, M.V Sednev, T. Lence, J.-Y. Roignant, J. König, C. Höbartner, Y. Motorin, A. Hildebrandt, M. Helm
NOseq: amplicon sequencing evaluation method for RNA m6A sites after chemical deamination
Nucleic Acids Res. 2021, 49, e23. First published 11 December 2020. doi: 10.1093/nar/gkaa1173 SI
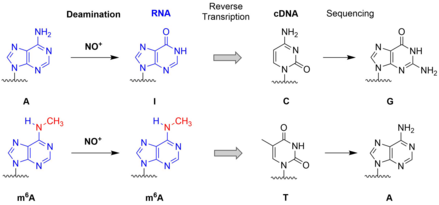
C.P.M. Scheitl, M. Ghaem Maghami, A.-K. Lenz, C. Höbartner
Site-specific RNA methylation by a methyltransferase ribozyme
Nature 2020, 587, 663–667 . First published 28 Oct 2020. doi: 10.1038/s41586-020-2854-z SI
R. Micura, C. Höbartner,
Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes
Chem. Soc. Rev. 2020, 49, 7331-7353. First published 18 Sep 2020. doi: doi.org/10.1039/D0CS00617C
C.P.M. Scheitl, S. Lange, C. Höbartner
New Deoxyribozymes for the Native Ligation of RNA
Molecules 2020, 25, 3650. doi: doi.org/10.3390/molecules25163650 SI
A. Liaqat, C. Stiller, M. Michel, M.V. Sednev, C. Höbartner
N6-isopentenyladenosine in RNA determines the cleavage site of endonuclease deoxyribozymes
Angew. Chem. Int. Ed. 2020, 59, 18627-18631. First published 18 July 2020. doi: https://doi.org/10.1002/anie.202006218 SI
M. Ghaem Maghami, S. Dey, A.-K. Lenz, C. Höbartner,
Repurposing antiviral drugs for orthogonal RNA‐catalyzed labeling of RNA
Angew. Chem. Int. Ed. 2020, 59, 9335–9339. Angew. Chem. 2020, 132, 9421-9425. First published 11 March 2020. doi: https://doi.org/10.1002/anie.202001300 SI
S. Werner, L. Schmidt, V. Marchand, T. Kemmer, C. Falschlunger, M.V. Sednev, G. Bec, E. Ennifar, C. Höbartner, R. Micura, Y. Motorin, A. Hildebrandt, M. Helm,Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes
Nucleic Acids Res. 2020, 48, 3734-3746. First published 25 February 2020. doi: https://doi.org/10.1093/nar/gkaa113 SI
C. Steinmetzger, C. Bäuerlein, C. Höbartner,
Supramolecular fluorescence resonance energy transfer in nucleobase‐modified fluorogenic RNA aptamers
Angew. Chem. Int. Ed. 2020, 59, 6760-6764. Angew. Chem. 2020, 132, 6826-6830. First published 12 February 2020.
doi: https://doi.org/10.1002/anie.201916707 SI
2019
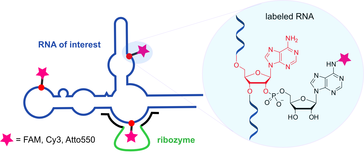
M. Ghaem Maghami, C.P.M. Scheitl, C. Höbartner,Direct in vitro selection of trans-acting ribozymes for posttranscriptional, site-specific, and covalent fluorescent labeling of RNA
J. Am. Chem. Soc. 2019, 141, 19546-19549. doi: https://pubs.acs.org/doi/10.1021/jacs.9b10531
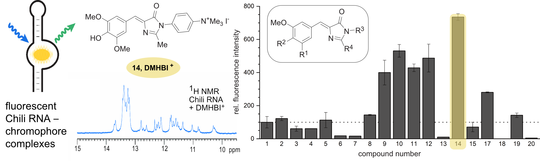
C. Steinmetzger, I. Bessi, A.-K. Lenz, C. Höbartner,
Structure–fluorescence activation relationships of a large Stokes shift fluorogenic RNA aptamer
Nucleic Acids Res. 2019, 47, 11538-11550. doi.org/10.1093/nar/gkz1084 SI
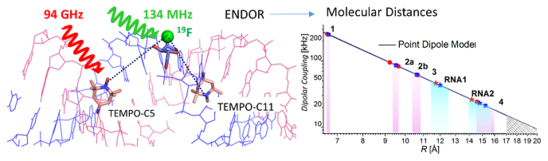
A. Meyer, S. Dechert, S. Dey, C. Höbartner, M. Bennati,Measurement of Angstrom to Nanometer Molecular Distances with 19F Nuclear Spins by EPR/ENDOR Spectroscopy
Angew. Chem. Int. Ed. 2020, 58, 373-379. Angew. Chem. 2020, 132, 381-387. First published 20 September 2019. doi.org/10.1002/anie.201908584
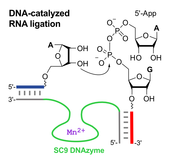
C. Höbartner, How DNA catalyzes RNA ligation
Nature Catalysis 2019, 2, 483–484. doi.org/10.1038/s41929-019-0295-6
News&View to: doi.org/10.1038/s41929-019-0290-y
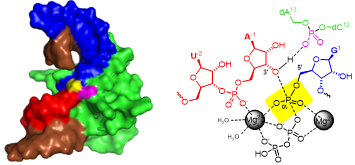
C. Steinmetzger, N. Palanisamy, K.R. Gore, C. Höbartner,
A multicolor large Stokes shift fluorogen‐activating RNA aptamer with cationic chromophores
Chem. Eur. J. 2019, 25, 1931-1935. doi.org/10.1002/chem.201805882. SI
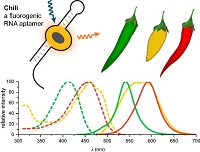
K.E. Bohnsack, C. Höbartner, M.T. Bohnsack,
Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease
Genes 2019, 10(2), 102. doi: 10.3390/genes10020102
2018
T.P. Hoernes, K. Faserl, M.A. Juen, J. Kremser, C. Gasser, E. Fuchs, X. Shi, A. Siewert, H. Lindner, C. Kreutz, R. Micura, S. Joseph, C. Höbartner, E. Westhof, A. Hüttenhofer, M.D. Erlacher,
Translation of non-standard codon nucleotides reveals minimal requirements for codon-anticodon interactions
Nat. Commun. 2018 Nov 19;9(1):4865. doi: 10.1038/s41467-018-07321-8. SI
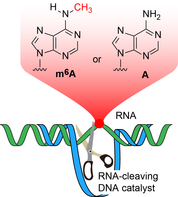
M.V. Sednev, V. Mykhailiuk, P. Choudhury, J. Halang, K.E. Sloan, M.T. Bohnsack, C. Höbartner,
N6‐Methyladenosine‐Sensitive RNA‐Cleaving Deoxyribozymes
Angew. Chem. Int. Ed. 2018, 57, 15117-15121. (VIP) Angew. Chem. 2018, 130, 15337-15341 SI
B. Moro, U. Chorostecki, S. Arikit, I.P. Suarez, C. Höbartner, R.M. Rasia, B.C. Meyers, J.F. Palatnik, Efficiency and precision of microRNA biogenesis modes in plants
Nucleic Acids Res. 2018, 46, 10709–10723. SI
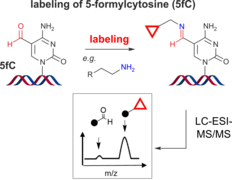
J. Dietzsch, D. Feineis, C. Höbartner, Chemoselective labeling and site-specific mapping of 5-formylcytosine as a cellular nucleic acid modification
FEBS Lett. 2018, 592, 2032-2047.
J. Kretschmer, H. Rao, P. Hackert, K.E. Sloan, C. Höbartner, M.T. Bohnsack,
The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1.
RNA 2018, 24, 1339-1350.
2017
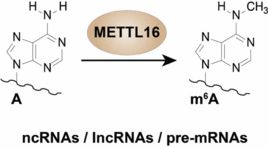
A.S. Warda, J. Kretschmer, P. Hackert, C. Lenz, H. Urlaub, C. Höbartner, K.E. Sloan, M.T. Bohnsack,Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs.
EMBO Reports 2017, 18, 2004-2014. SI
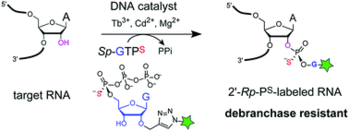
T. J. Carrocci, L. Lohe, M. J. Ashton, C. Höbartner and A. Hoskins,
Debranchase-resistant labeling of RNA using the 10DM24 deoxyribozyme and fluorescent modified nucleotides.
Chem. Commun. 2017, 53, 11992-11995. SI
M.A. Gomes de Castro, C. Höbartner, F. Opazo,
Staining of Membrane Receptors with Fluorescently-labeled DNA Aptamers for Super-resolution Imaging.
Bio-protocol 7(17): e2541.
P. Bao, C. Höbartner, K. Hartmuth, R. Lührmann,
Yeast Prp2 liberates the 5' splice site and the branch site adenosine for catalysis of pre-mRNA splicing.
RNA 2017, 1770-1779.
S. Haag, K.E. Sloan, C. Höbartner, M.T. Bohnsack, In Vitro Assays for RNA Methyltransferase Activity.
Methods Mol. Biol. 2017, 1562 : 259-268.
M.A. Gomes de Castro, C. Höbartner, F. Opazo, Aptamers provide superior stainings of cellular receptors studied under super-resolution microscopy.
PLoS One 2017, 12, e0173050.
K.E. Sloan, C. Höbartner, M.T. Bohnsack, How RNA modification allows nonconventional decoding in mitochondria.
Cell Cycle 2017, 16, 145-146. (invited Feature)
2016
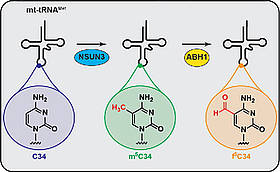
S. Haag, K.E. Sloan, N. Ranjan, A.S. Warda, J. Kretschmer, C. Blessing, B. Hübner, J. Seikowski, S. Dennerlein, P. Rehling, M.V. Rodnina, C. Höbartner, M.T. Bohnsack,
NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation
EMBO Journal 2016, 35, 2104-2119. SI
comments by others:
F. Boos, M. Wollin, J.M. Herrmann,
Methionine on the rise: how mitochondria changed their codon usage
News & Views EMBO J. 2016 Aug 30.
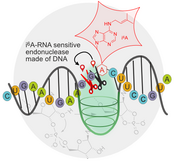
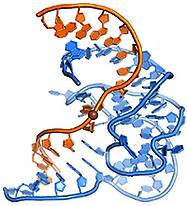
A. Ponce-Salvatierra, K. Wawrzyniak-Turek, U. Steuerwald, C. Höbartner, V. Pena, Crystal structure of a DNA catalyst.
Nature 2016, 529, 231-234.
comments by others:
K. Woolcock, Structure of a DNA enzyme.Nat. Struct. Mol. Biol.2016, 23, 97.
T.L. Sheppard, Catalytic DNA: Ligating with lambda.Nat. Chem. Biol.2016, 12, 125.
M. Kostic, Select: Structure of a DNAzyme.Cell Chem Biol.2016 Feb 18;23(2):200.
J. Wirmer-Bartoschek, H. Schwalbe, Understanding How DNA Enzymes Work.
Angew. Chem. Int. Ed. 2016, 55, 5376.
Press release: MPIbpc, Univ. Göttingen
F1000Prime Recommendations by S.K. Silverman; D. Wetmore; B. Seelig; I. Hirao;
total score: 11.
Scientists Report First Crystal Structure Of A DNAzyme C&E News
M. Theuser, C. Höbartner, M.C. Wahl, K.F. Santos, Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase.
Proc. Natl. Acad. Sci. USA. 2016, 113, 7798-7803. SI
B. Samanta, J. Seikowski, C. Höbartner, Fluorogenic labeling of 5-formylpyrimidines in DNA and RNA.
Angew. Chem. Int. Ed. 2016, 55, 1912-1916. Angew. Chem. 2016, 130, 15337-15341. SI
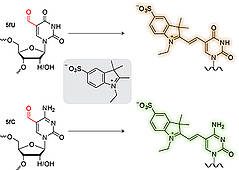
F. Javadi-Zarnaghi, C. Höbartner, Functional hallmarks of a catalytic DNA that makes lariat RNA.
Chem. Eur. J. 2016, 22, 3720-3726. SI
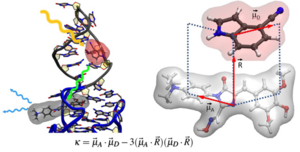
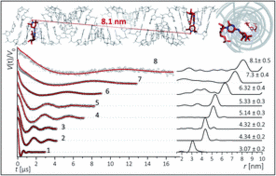
K. Halbmair, J. Seikowski, I. Tkach, C. Höbartner, D. Sezer, M. Bennati, High-resolution measurement of long-range distances in RNA: pulse EPR spectroscopy with TEMPO-labeled nucleotides.Chem. Sci. 2016, 7, 3172-3180. SI
S. Kwiatkowski, V.M. Sviripa, Z. Zhang, A.E. Wendlandt, C. Höbartner, D.S. Watt*, S. Stamm*, Synthesis of a norcantharidin-tethered guanosine: Protein phosphatase-1 inhibitors that change alternative splicing.
Bioorg. Med. Chem. Lett. 2016, 26, 965-968. SI (* co-corresponding author)
2015
M. Kosutic, S. Neuner, A. Ren, S. Flür, C. Wunderlich, E. Mairhofer, N. Vusurovic, J. Seikowski, K. Breuker, C. Höbartner, D.J. Patel, C. Kreutz, R. Micura, A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew. Chem. Int. Ed. 2015, 54, 15128-15133. Angew. Chem. 2015, 127, 15343-15348. SI
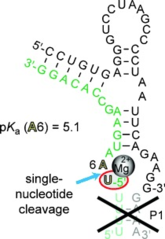
S. Haag, A.S. Warda, J. Kretschmer, M.A. Günnigmann, C. Höbartner, M.T. Bohnsack, NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs.
RNA 2015, 21, 1532-1543. SI
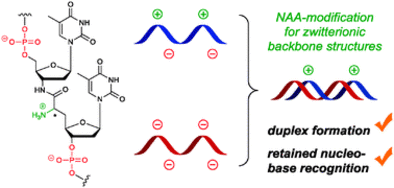
B. Schmidtgall, C. Höbartner, C. Ducho, NAA-modified DNA oligonucleotides with zwitterionic backbones: stereoselective synthesis of A-T phosphoramidite building blocks.Beilstein J. Org. Chem. 2015, 11, 50-60. SI

E. Turriani, C. Höbartner, T.M. Jovin, Mg2+-dependent conformational changes and product release during DNA-catalyzed RNA ligation monitored by Bimane fluorescence.Nucleic Acids Res. 2015, 43, 40-50. SI
Z. Warkocki, C. Schneider, S. Mozaffari-Jovin, J. Schmitzová, C. Höbartner, P. Fabrizio, R. Lührmann, The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome.Genes Dev. 2015, 29, 94-107. SI
2014
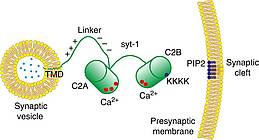
C.C. Lin, J. Seikowski, A. Pérez-Lara, R. Jahn, C. Höbartner, P.J. Walla, Control of membrane gaps by synaptotagmin-Ca2+ measured with a novel membrane distance ruler.
Nat. Commun. 2014 Dec 15; 5, 5859. SI
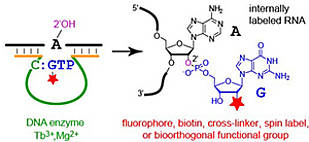
L. Büttner, F. Javadi-Zarnaghi, C. Höbartner, Site-specific labeling of RNA at internal ribose hydroxyl groups: terbium-assisted deoxyribozymes at work.
J. Am. Chem. Soc. 2014, 136, 8131-8137. SI
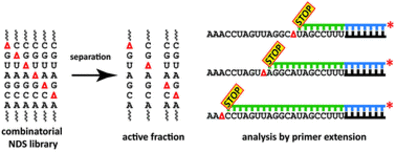
K. Wawrzyniak-Turek, C. Höbartner, Enzymatic combinatorial nucleoside deletion scanning mutagenesis of functional RNA.
Chem. Commun. 2014, 50, 10937-10940. SI
K. Wawrzyniak-Turek, C. Höbartner, Deoxyribozyme-mediated ligation for incorporating EPR spin labels and reporter groups into RNA.
Methods Enzymol. 2014, 549, 85-104.
B. Schmidtgall, F. Wachowius, C. Höbartner, C. Ducho, Synthesis and properties of DNA oligonucleotides with a zwitterionic backbone structure.
Chem. Commun. 2014, 50, 13742-13745. SI
I. Tkach, K. Halbmair, C. Höbartner, M. Bennati, High-frequency 263 GHz PELDOR. Appl.
Magn. Res. 2014, 45, 969-979.
2013
F. Javadi-Zarnaghi, C. Höbartner, Lanthanide cofactors accelerate DNA-catalyzed synthesis of branched RNA.
J. Am. Chem. Soc. 2013, 135, 12839-12848. SI
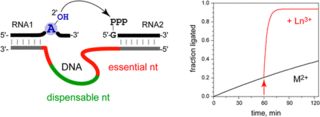
L. Büttner, J. Seikowski, K. Wawrzyniak, A. Ochmann, C. Höbartner, Synthesis of spin-labeled riboswitch RNAs using convertible nucleosides and DNA-catalyzed RNA ligation.
Bioorg. Med. Chem. 2013, 21, 6171-6180. SI
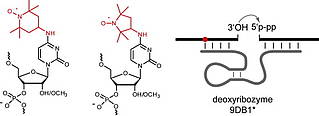
B. Samanta, C. Höbartner, Combinatorial Nucleoside-Deletion-Scanning Mutagenesis of Functional DNA.
Angew. Chem. Int. Ed. 2013, 52, 2995-2999. Angew. Chem.2013, 125, 3069-3073. SI
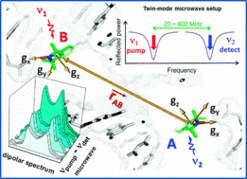
I. Tkach, S. Pornsuwan, C. Höbartner, F. Wachowius, S.T. Sigurdsson, T. Baranova, U. Diederichsen, G. Sicoli, M. Bennati, Orientation selection in distance measurements between nitroxide spin labels at 94 GHz EPR with variable dual frequency irradiation.Phys. Chem. Chem. Phys. 2013, 15, 3433-3437. SI
2012
C. Höbartner*, G. Sicoli, F. Wachowius, D.B. Gophane, S.T. Sigurdsson*, Synthesis and Characterization of RNA Containing a Rigid and Nonperturbing Cytidine-Derived Spin Label.
J. Org. Chem. 2012, 77, 7749-7754. SI (* co-corresponding authors)
G.N. Nawale, K.R. Gore, C. Höbartner*, P.I. Pradeepkumar*, Incorporation of 4'-C-aminomethyl-2'-O-methylthymidine into DNA by thermophilic DNA polymerases.
Chem. Commun. 2012, 48, 9619-9621. (* co-corresponding authors)
K.R. Gore, G.N. Nawale, S. Harikrishna, V. G. Chittoor, S.K. Pandey, C. Höbartner, S. Patankar, P.I. Pradeepkumar, Synthesis, Gene Silencing, and Molecular Modeling Studies of 4'-C-Amino-methyl-2'-O-methyl Modified Small Interfering RNAs.J. Org. Chem. 2012, 77, 3233-3245. SI
2011
F. Wachowius, C. Höbartner, Probing essential nucleobase functional groups in aptamers and deoxyribozymes by nucleotide analog interference mapping of DNA.J. Am. Chem. Soc. 2011, 133, 14888-14891. SI
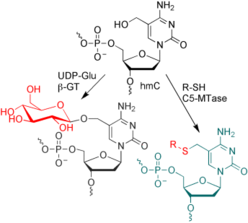
C. Höbartner, Enzymatic labeling of 5-hydroxymethylcytosine in DNA.
Angew. Chem. Int. Ed. 2011, 50, 4268-4270. Angew. Chem.2011, 123, 4357-4359. (Highlight)
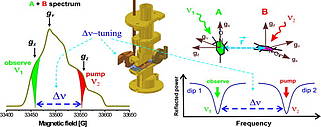
I. Tkach, G. Sicoli, C. Höbartner and M. Bennati, A dual-mode microwave resonator for double electron-electron spin resonance spectroscopy at W-band microwave frequencies.J. Mag. Res. 2011, 209, 341-346.
N. Dixit, R.K. Koiri, B.K. Maurya, S.K. Trigun, C. Höbartner, L. Mishra, One pot synthesis of Cu(II) 2,2'-bipyridyl complexes of 5-hydroxy-hydurilic acid and alloxanic acid: Synthesis, crystal structure, chemical nuclease activity and cytotoxicity.J. Inorg.Biochem. 2011, 105, 256-267.
2010
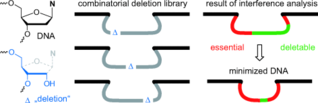
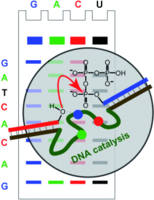
F. Wachowius, F. Javadi-Zarnaghi, C. Höbartner, Combinatorial Mutation Interference Analysis reveals functional nucleotides required for DNA catalysis.
Angew. Chem. Int. Ed. 2010, 49, 8504-8508. Angew. Chem.2010, 122, 8682-8687. SI
comments by others
comment in Faculty of 1000 Biology: Müller S.: 2010.
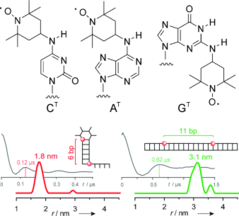
G. Sicoli, F. Wachowius, M. Bennati, C. Höbartner*, Secondary Structure Probing of Spin-labeled RNA by Pulsed EPR Spectroscopy.
Angew. Chem. Int. Ed. 2010, 49, 6443-6447. Angew. Chem. 2010, 122, 6588-6592.
SI (* co-corresponding author)
F. Wachowius, C. Höbartner, Chemical RNA modifications for studies of RNA structure and dynamics.
ChemBioChem 2010, 11, 469-480.
2009
D.A. Heller, H. Jin, B.M. Martinez, D. Patel, B.M. Miller, T.-K. Yeung, P.V. Jena, C. Höbartner, T. Ha, S.K. Silverman, M.S. Strano, Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes.
Nat. Nanotechnol. 2009, 4, 114-120. SI
comments by others
News & views: T.D. Krauss, Nat. Nanotechnol.2009, 4, 85-86.
R. Rieder, C. Höbartner, R. Micura, Enzymatic ligation strategies for the preparation of purine riboswitches with site-specific chemical modifications.
in Riboswitches (Ed. A. Serganov), Methods Mol. Biol. 2009, 540, 15-24.
2008
P. I. Pradeepkumar, C. Höbartner, D.A. Baum S. K. Silverman, DNA-catalyzed formation of nucleopeptide linkages.
Angew. Chem. Int. Ed. 2008, 47, 1753-1757. Angew. Chem. 2008, 120, 1777-1781. SI
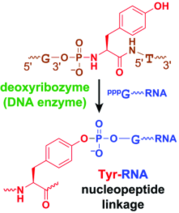
2007
C. Höbartner, S. K. Silverman, Recent Advances in DNA Catalysis.
Biopolymers 2007, 87, 279-292.
C. Höbartner, S. K. Silverman, Engineering a Selective Small-Molecule Substrate Binding Site into a Deoxyribozyme.
Angew. Chem. Int. Ed. 2007, 46, 7420-7424. Angew. Chem. 2007, 119, 7564-7568. SI
C. Höbartner, P. I. Pradeepkumar, S. K. Silverman, Site-Selective Depurination by a Periodate-Dependent Deoxyribozyme.
Chem. Commun. 2007, 2255-2257.

2006
R. Micura, C. Höbartner, R. Rieder, C. Kreutz, B. Puffer, K. Lang, H. Moroder, Preparation of 2'-Deoxy-2'-Methylseleno-Modified RNA.
Current Protocols in Nucleic Acid Chemistry 2006, 1.15.1-1.15.34.
2005
C. Höbartner, S. K. Silverman, Modulation of RNA tertiary folding by incorporation of caged nucleotides.
Angew. Chem. 2005, 117, 7471-7475; Angew. Chem. Int. Ed. 2005, 44, 7305-7309. SI
C. Höbartner, R. Rieder, C. Kreutz, B. Puffer, K. Lang, A. Polonskaia, A. Serganov, R. Micura, Syntheses of RNAs with up to 100 nucleotides containing site-specific 2'-Se-methyl labels for use in X-ray crystallography.
J. Am. Chem. Soc. 2005, 127, 12035-12045.
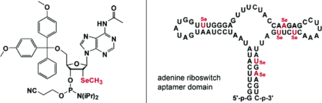
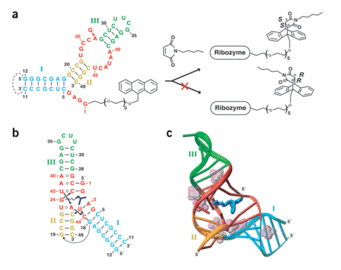
A. Serganov, S. Keiper, L. Malinina, V. Tereshko, E. Skripkin, C. Höbartner, A. Polonskaia, A. T. Phan, R. Wombacher, R. Micura, Z. Dauter, A. Jäschke, D. J. Patel, Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat. Struct. Mol. Biol. 2005, 12, 218-224. SI
comments by others
News and views: JN Pitt, A. Ferré-d'Amare, Nat. Struct. Mol. Biol. 2005, 12, 206-208.
2004
A. Serganov, Y. R. Yuan, O. Pikovskaya, A. Polonskaia, L. Malinina, A.T. Phan, C. Höbartner, R. Micura, R.R. Breaker, D.J. Patel, Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs.
Chem. Biol. 2004, 11, 1729-1741.
comments by others
Comment: A. Lescoute, E. Westhof, Chem. Biol. 2005, 12, 10-13.
C. Höbartner, H. Mittendorfer, K. Breuker, R. Micura, Triggering of RNA secondary structures by a functionalized nucleobase.
Angew. Chem. 2004, 116, 4012-4015; Angew. Chem. Int. Ed. 2004, 43, 3922-3925. SI
C. Höbartner, R. Micura, Chemical synthesis of selenium-modified oligoribonucleotides and their enzymatic ligation leading to an U6 snRNA stem-loop segment. J. Am. Chem. Soc. 2004, 126, 1141-1149. SI
2003
R. Micura and C. Höbartner, On secondary structure rearrangements and equilibria of small RNAs.
ChemBioChem 2003, 4, 984-990.
C. Höbartner, C. Kreutz, E. Flecker, E. Ottenschläger, W. Pils, K. Grubmayr, R. Micura, The synthesis of 2'-O -[(triisopropylsilyl)oxy]methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m22G, m1I, m3U, m4C, m6A, m62A) for use in automated RNA solid-phase synthesis.Monatshefte für Chemie - Chemical Monthly 2003, 134, 851-873.
C. Höbartner, R. Micura, Bistable secondary structures of small RNAs and their structural probing by comparative imino proton NMR spectroscopy.
J. Mol. Biol. 2003, 325, 421-431.
2002
C. Höbartner, M.-O. Ebert, B. Jaun, R. Micura, RNA two-state conformation equilibria and the effect of nucleobase methylation.
Angew. Chem. 2002, 114, 619-623; Angew. Chem. Int. Ed. 2002, 41, 605-609.
2001
R. Micura, W. Pils, C. Höbartner, K. Grubmayr, M.-O. Ebert, B. Jaun, Methylation of nucleobases in RNA oligonucleotides mediates hairpin-duplex conversion.
Nucleic Acids Res. 2001, 29, 3977-4005.
2000
A. Stutz, C. Höbartner, S. Pitsch, Novel fluoride-labile nucleobase-protecting groups for the synthesis of 3'(2')-O -aminoacylated RNA sequences.
Helv. Chim. Acta 2000, 83, 2477-2503.
Buchkapitel
A. Liaqat, M.V. Sednev, C. Höbartner, In vitro Selection of Deoxyribozymes for the Detection of RNA Modifications. In: Ribosome Biogenesis: Reviews, Methods and Protocols. Springer 2021, 167–179. ISBN: 978-1-0716-2501-9_10
F. Javadi-Zarnaghi, C. Höbartner, Strategies for Characterization of Enzymatic Nucleic Acids. In: Advances in Biochemical Engineering/Biotechnology. Springer, Berlin, Heidelberg 2017, 1-22. ISBN: 978-3-030-29646-9
C. Höbartner, L. Büttner, F. Javadi-Zarnaghi, Bioorthogonal modifications and cycloaddition reactions for RNA Chemical Biology, in RNA Structure and Folding (Eds. D. Klostermeier, C. Hammann). DeGruyter 2013, 75-99. ISBN 978-3110284591
P.I. Pradeepkumar, C. Höbartner, RNA-Cleaving DNA enzymes and their potential therapeutic applications as antibacterial and antiviral agents, in From Nucleic Acids Sequences to Molecular Medicine (Eds. V. Erdmann, J. Barciszewski), Springer, 2012, 371-410. ISBN 978-3-642-27425-1
C. Höbartner, P.I. Pradeepkumar, DNA catalysts for synthetic applications in biomolecular chemistry, in New strategies in chemical synthesis and catalysis (Ed. B. Pignataro), Wiley-VCH, 2012, 129-155. ISBN 978-3-527-33090-4
C. Höbartner, Chemical Synthesis of RNA, in Alternative pre-mRNA splicing: Theory and Protocols (Eds. S. Stamm, C. Smith, R. Lührmann), Wiley-VCH, 2012, 155-163. ISBN 3-527-32606-5
C. Höbartner, F. Wachowius, Chemical synthesis of modified RNA, in The Chemical Biology of Nucleic Acids (Ed. G. Mayer), Wiley, 2010, 1-37. ISBN: 978-0-470-51974-5
R. Micura and C. Höbartner, Equilibria of RNA secondary structures, in Highlights in Bioorganic Chemistry, Methods and Applications (Ed. C. Schmuck, H. Wennemers) Wiley-VCH, Weinheim, 2004, 3-16.