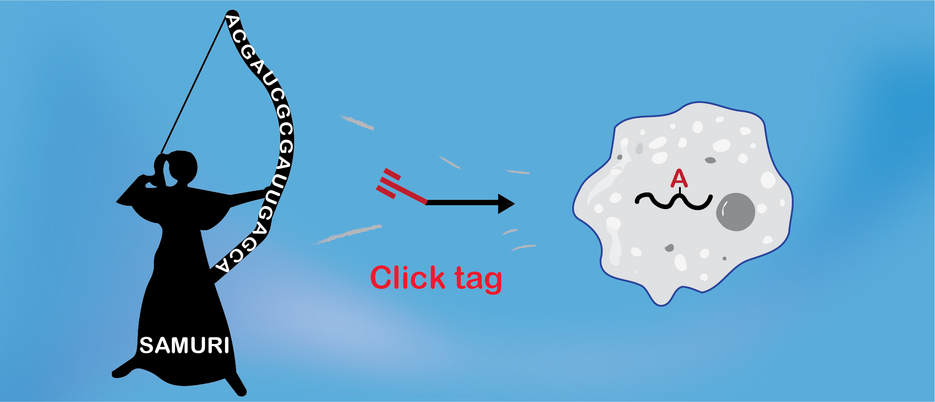
SAMURI targets a specific adenosine in RNA
06.09.2023We report the ribozyme SAMURI which uses a synthetic SAM analogue as cofactor to make RNA in cells accessible for labeling by Click Chemistry. Just published in Nature Chemistry. Congrats to Takumi Okuda and all coauthors!
A new ribozyme for RNA labeling and exploring the world of RNA
The molecular composition of RNA as a linear biopolymer is rather simple, but the structural and functional diversity of RNA is still full of surprises. Defined three-dimensional structures are the basis for RNA’s many roles in biology and enable new functions, for example, to specifically recognize other molecules and to catalyze chemical reactions. Such RNA catalysts are called ribozymes.
The team of chemistry professor Claudia Höbartner at the Julius-Maximilians-Universität Würzburg now reports the discovery of a new ribozyme in the journal Nature chemistry. The ribozyme was nicknamed “SAMURI”, which stands for SAM analogue utilizing ribozyme. SAM (S-Adenosylmethionine) is the most important methyl group donor in biology. The ribozyme was found in a large pool of synthetic RNA and catalyzes the site-specific alkylation of a target RNA.
Exploring the ability of RNA to catalyze the installation of RNA modifications, such as methyl groups, is of fundamental interest in the field of RNA chemistry. The Höbartner group recently reported the first methyltransferase ribozyme MTR1 that uses a guanine cofactor. Now, they turned their attention to SAM which is naturally used by protein enzymes. Höbartner and her team wondered if this ancient cofactor could be used by ribozymes, and if a new ribozyme-cofactor combination could be repurposed for RNA labeling.
Takumi Okuda, a postdoctoral researcher in the Höbartner lab designed and synthesized a chemically modified SAM analogue that was suitable for finding the new ribozyme. He explains: “We need to balance the stability and the reactivity of the cofactor to allow the transfer of an alkyne group for enrichment by click chemistry.”
SAMURI was shown to recognize its target RNA by Watson-Crick base pairing and can be engineered for diverse target sites. Importantly, SAMURI is active under physiological conditions and was shown to be active in cells.
“Ribozymes are useful as RNA-based tools that contribute insights into RNA functions and will allow us to visualize RNAs directly, and thereby help to explore the localization, transport and interactions of RNA with other molecules in the cell,” says Takumi Okuda.
Being able to install native RNA modifications by ribozymes instead of protein enzymes is of fundamental interest for understanding the evolution of functional RNAs and the functional roles of posttranscriptional modifications. “In addition, we foresee new applications of ribozymes to rescue RNA modification states when the responsible proteins are not available or dysfunctional, for example, due to mutations,” adds Claudia Höbartner.
The results reported in this study encourage the search for additional ribozymes and promise a range of applications in the fields of RNA chemical biology and RNA medicine.
Read the full story about SAMURI in Nature Chemistry: A SAM-analogue-utilizing ribozyme for site-specific RNA labeling in living cells.
Press release by the University of Würzburg