Nanocosmos of Cells Under the Magnifying Glass
09/17/2014Jürgen Seibel, biobiophysicist Markus Sauer and their coworkers have managed to take a unique look at the membranes of human cells using a new technique. This technique that they have devised makes individual saccharified proteins and lipids visible at the molecular level.


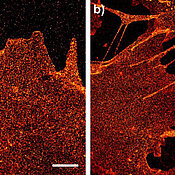
Picture a satellite orbiting the Earth and capturing numerous individual images of the planet at night in impressive definition, which, when combined to create one single large photograph, deliver an extremely detailed picture of life at night on the surface of the Earth. Only, in this case, the camera is not orbiting the Earth; it is traveling across human cells. And the light captured does not come from street lights, headlights, and lamps, but from specific structures inside the cell membrane or, more accurately, from glycoproteins and lipids illuminated using a special technique.
The people responsible for these images are Professor Markus Sauer and Professor Jürgen Seibel, a biophysicist and chemist, respectively, from the University of Würzburg. They report on their new findings in the renowned journal “Angewandte Chemie” [Applied Chemistry]; the publishers have even rated their work as a “hot paper”.
Modified sugar molecules and luminescent dyes
“We created sugar-like structures chemically and administered them to the nutrient solution of human cells,” is how Jürgen Seibel explains the procedure the chemists followed. The cells metabolized these molecules and integrated them into their membranes biosynthetically on their surface. The trick: “We modified the sugar molecules slightly so they can be combined with a fluorescent dye,” says Seibel.
The biophysicists’ job was then to illuminate these dyes in a suitable manner so that an image could be generated of the individual molecules on the cell membrane. The technique used was developed by Markus Sauer and his team. Its name: dSTORM – direct Stochastic Optical Reconstruction Microscopy. This is a specific form of high-resolution fluorescence microscopy; it allows images to be captured of cellular structures and molecules at a resolution that is ten times better than ever before. The size of the objects displayed is between 20 and 30 nanometers, i.e. millionths of millimeters.
An Off switch stops the fluorescence
“dSTORM microscopy uses commercial fluorescent dyes which, when exposed to light of a suitable wavelength in the presence of thiols, transition to a reduced and very stable optical ‘off’ state,” says Markus Sauer to explain the principle behind this technique. To put it another way: the dyes stop fluorescing for a few seconds.
It may sound strange at first to deactivate dyes so that a high-resolution image can be generated, but this makes sense when you know the other details: “Once the cell has been exposed, more than 99.9 percent of the dyes are quickly deactivated. But a few continue to shine,” says the biophysicist. The scientists are able to spatially distinguish their signals and thus calculate the exact position of the dye. It is therefore possible to localize individual dye molecules. After that, these molecules, too, lapse into the inactive ‘off’ state.
The scientists repeat this process numerous times and then piece the many “individual images” together to create one overall image. “According to the rule of stochastic randomness, all the dyes are returned to their fluorescent ‘on’ state and localized individually,” says Sauer. A finished image is created once all the molecules have emitted their signals.
Precise statements about location and quantity
The two scientists have counted up to 1600 glycoproteins and glycolipids per square micrometer on the surface of human cells using this method. This means that a single cell carries some five million of these building blocks in total. Humans are made up of around a trillion cells.
The studies conducted at the University of Würzburg are making it possible to localize and quantify the number of sugars on cell surfaces exactly for the very first time. This is of particular interest in the research into infectious diseases and cancer, explains Jürgen Seibel. This is because macromolecules containing carbohydrates, known as glycoproteins and glycolipids, control immune responses, cell growth, and cell death on the cell surface.
Tumors and bacteria, and also viruses, imitate and exploit the natural recognition process and infect human cells. The Würzburg scientists are hoping that their new method will provide deeper insight into such biological happenings. Their work was performed as part of the “3D Super-Resolution” collaborative project, which is funded by the Federal Ministry of Education and Research.
„Super-Resolution Imaging of Plasma Membrane Glycans”, Sebastian Letschert, Antonia Göhler, Christian Franke, Nadja Bertleff-Zieschang, Elisabeth Memmel, Sören Doose, Jürgen Seibel, Markus Sauer, Angewandte Chemie, online publiziert am 22. August 2014, DOI: 10.1002/ange.201406045
Contact
Prof. Dr. Jürgen Seibel, T +49 931 31-85326, seibel@chemie.uni-wuerzburg.de